FUNCTIONAL ANNOTATION OF PROTEINS
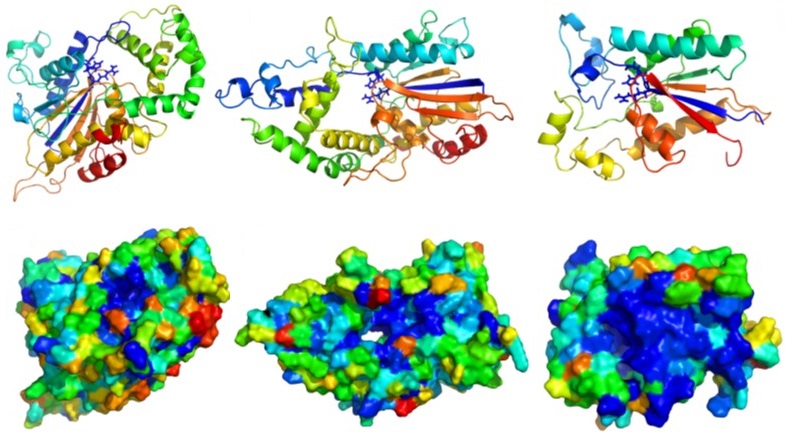

My MRes in Post-Genomic Science project involved using various bioinformatics techniques to functionally annotate members of the histidine phosphatase superfamily, including the collection of sequences from public databases using the iterative search program “Jackhmmer”, detecting “clusters” using CLANS for a “neural network based approach”, STRING to visualise genomic context and 3D homology modelling & small molecule docking using RosettaCM.
Grade: Distinction
The aim of the study was to identify novel functions for members of the Histidine Phosphatase Superfamily, focussing on large groups of hypothetical proteins. To achieve this, the first phase of the investigation was the collection of a complete set of Histidine Phosphatase sequences using the iterative search program Jackhmmer (Eddy, 1998; Finn et al., 2011). The second phase was to partition the collected sequences, using sequence similarity, into "clusters" that likely share the same function by utilising CLANS (Frickey & Lupas, 2004; Frickey & Weiller, 2007). As the emphasis was the prediction of novel functions, the next stage was to eliminate clusters of Histidine Phosphatases with known functions by performing database searches. The final phase was to predict the functions of the remaining large clusters of HPs, using a variety of bioinformatics methods including genomic context, homology modelling and metabolite docking. STRING (Szklarczyk et al., 2015) was proposed for visualising genomic context, identifying predicted functional partners and determining potential ligands for subsequent docking, using the RosettaLigand (Combs et al., 2013), into homology modelling created with RosettaCM (Combs et al., 2013).
An Integrated Approach to Function Annotation in the Histidine Phosphatase Superfamily